3. Data journey - bumblebee pollinators
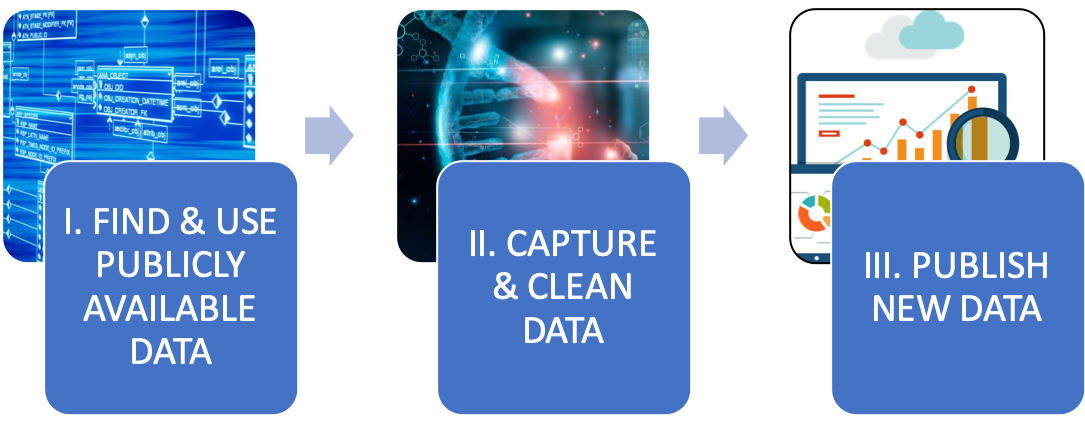
| This data journey is a series of practical exercises focusing on bumblebee pollinators. Using GBIF and BOLD, you will learn: I. how to find and use already available biodiversity data in connexion with your research questions; II. How to efficiently capture and clean this data – i.e. put them in a standard format that is directly relevant and exploitable for you; III. How to generate and publish new data according to a international standards. |
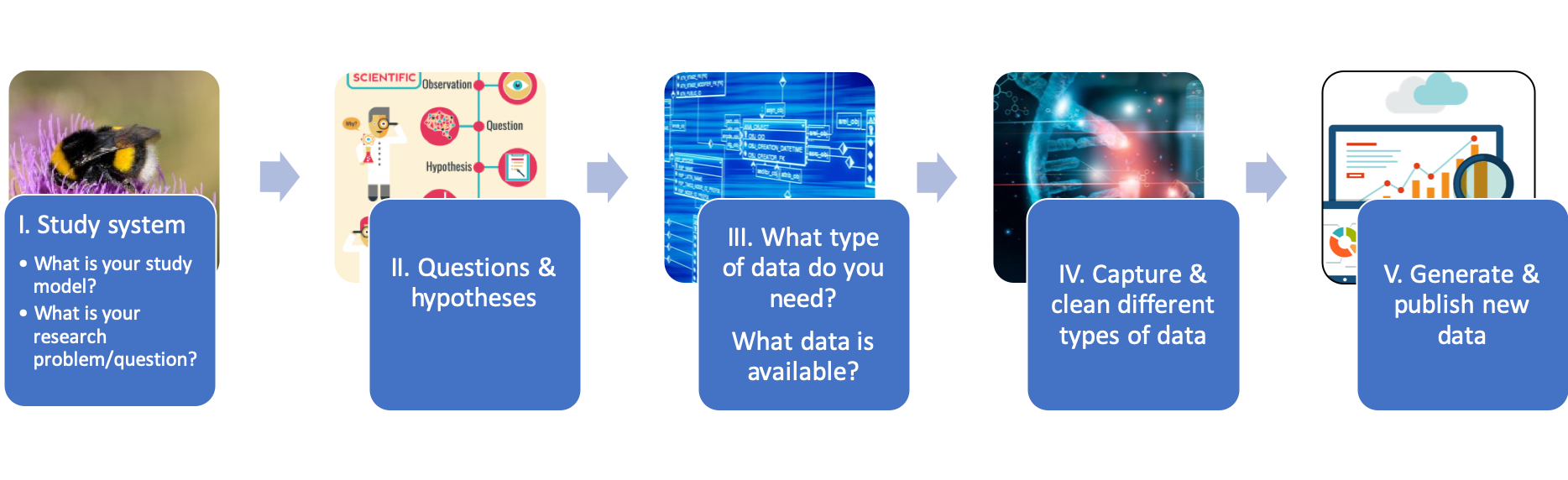
| This data journey is comprised of nine steps. Each step (or set of steps), correlate to the different modules of the course. After a series of theoretical lectures, you will return to the Data Journey to complete the practical exercises. The practical exercises follow a path: I. the study system; II. questions and hypotheses; III. availability of data; IV. capture and cleaning of data; and V. generate and publish data. |